A Bottom-Up Approach for Pancreas Segmentation Using Cascaded Superpixels and (Deep) Image Patch Labeling
Product Description
A Bottom-Up Approach for Pancreas Segmentation Using Cascaded Superpixels and (Deep) Image Patch Labeling
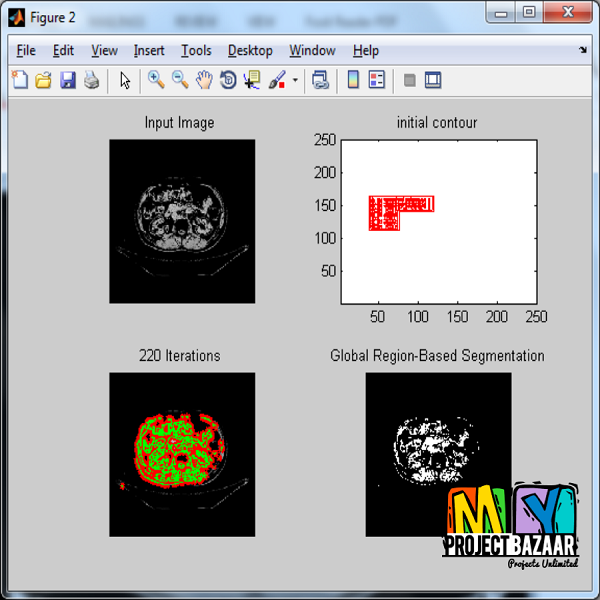
Abstract— Robust organ segmentation is a prerequisite for computer-aided diagnosis, quantitative imaging analysis, pathology detection, and surgical assistance. For organs with high anatomical variability (e.g., the pancreas), previous segmentation approaches report low accuracies, compared with well-studied organs, such as the liver or heart. We present an automated bottom-up approach for pancreas segmentation in abdominal computed tomography (CT) scans. The method generates a hierarchical cascade of information propagation by classifying image patches at different resolutions and cascading (segments) superpixels. The system contains four steps: 1) decomposition of CT slice images into a set of disjoint boundary-preserving super-pixels; 2) computation of pancreas class probability maps via dense patch labeling; 3) superpixel classification by pooling both intensity and probability features to form empirical statistics in cascaded random forest frameworks; and 4) simple connectivity based post-processing. Dense image patch labeling is conducted using two methods: efficient random forest classification on image histogram, location and texture features; and more expensive (but more accurate) deep convolutional neural network classification, on larger image windows. Over-segmented 2 − D CT slices by the simple linear iterative clustering approach are adopted through model/parameter calibration and labeled at the superpixel level for positive (pancreas)
or negative (non-pancreas or background) classes. < final year projects >
Including Packages
Our Specialization
Support Service
Statistical Report

satisfied customers
3,589
Freelance projects
983
sales on Site
11,021
developers
175+Additional Information
| Domains | |
|---|---|
| Programming Language |