Biclustering of human cancer micro array data using co-similarity based co-clustering
Product Description
Biclustering of human cancer micro array data using co-similarity based co-clustering
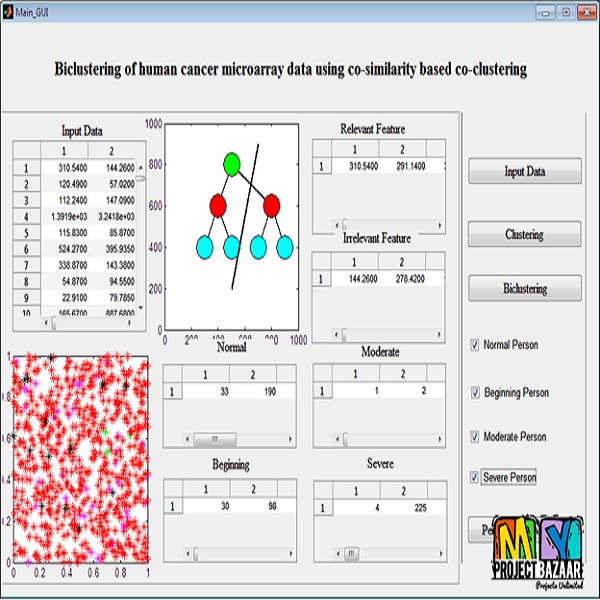
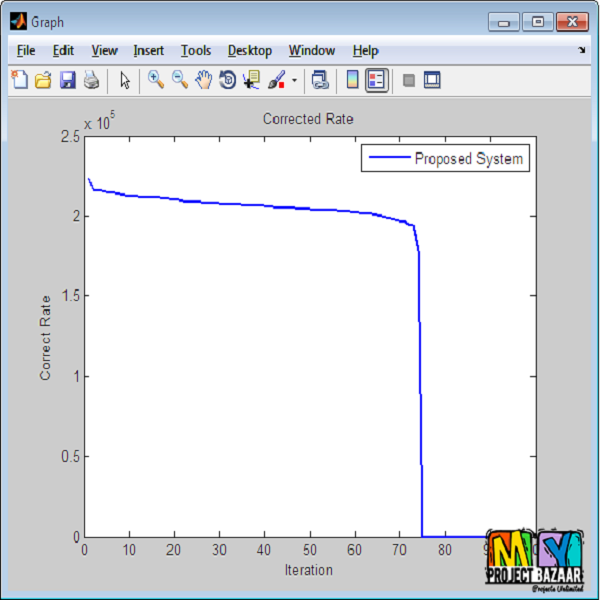
Abstract— Biclustering of gene expression data aims at finding localized patterns in a subspace. A bicluster (some-times called a co-cluster), in the context of gene expression data, is a set of genes that exhibit similar expression intensity under a subset of experimental features (conditions). Mostbiclustering algorithms proposed in the literature aim at finding sub-matrices that exhibit some sort of coherence by selecting an initial sub-matrix and iteratively adding or subtracting rows and columns. These algorithms are generally dependent on the initial, hard selection of the gene and condition clusters respectively. In this work, we adapt a recently proposed approach for clustering textual data to find biclusters in gene expression data. Our proposed technique is based on the concept of co-similarity between genes (and between conditions) that exploits weighted higher order paths in a bipartite graph representation of the gene expression data. Therefore, we build statistical relations between genes and between conditions by comparing all genes and conditions before finally extracting biclusters from the data. We show that the proposed technique is able to find meaningful non-overlapping biclusters both on synthetically generated data as well as real cancer data. Our results indicate that the proposed technique is resistant to noise in the data and can successfully retrieve biclusters even in the presence of relatively large amount of noise. We also analyse our results with respect to the discovered genes and observe that our extracted biclusters are supported by biological evidences, such as enrichment of gene functions and biological processes. < final year projects >
Including Packages
Our Specialization
Support Service
Statistical Report

satisfied customers
3,589
Freelance projects
983
sales on Site
11,021
developers
175+Additional Information
| Domains | |
|---|---|
| Programming Language |