
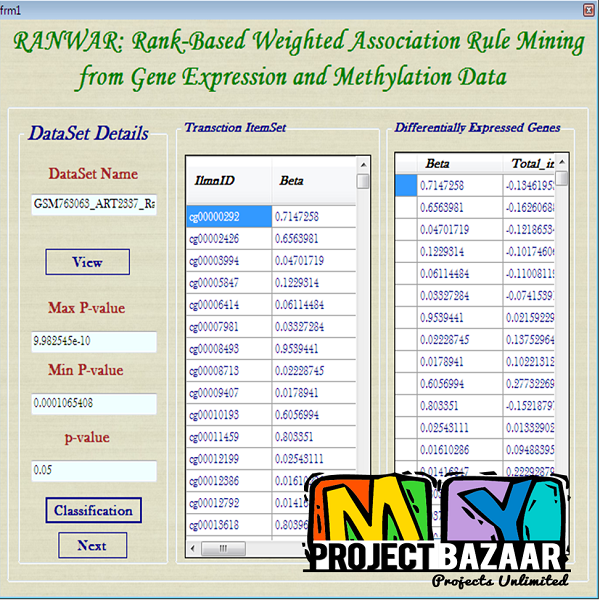
RANWAR: Rank-Based Weighted Association Rule Mining from Gene Expression and Methylation Data.
Product Description
RANWAR: Rank-Based Weighted Association Rule Mining from Gene Expression and Methylation Data
Abstract— RANWAR: Rank-Based Weighted Association Rule Mining from Gene Expression and Methylation Data. Ranking of association rules is currently an interesting topic in data mining and bioinformatics. The huge number of evolved rules of items (or, genes) by association rule mining (ARM) algorithms makes confusion to the decision maker. In this article, we propose a weighted rule-mining technique (say,RANWARor rank-based weighted association rule-mining) to rank the rules using two novel rule-interestingness measures, viz., rank-based weighted condensed support (wcs) and weighted condensed con-fidence < Final Year Projects 2016 > wcc measures to bypass the problem.These measures are basically depended on the rank of items (genes). Using the rank, we assign weight to each item. RANWAR generates much less number of frequent itemsets than the state-of-the-art association rule mining algorithms. Thus, it saves time of execution of the algorithm. We run RANWAR on gene expression and methylation datasets. The genes of the top rules are biologically validated by Gene Ontologies (GOs) and KEGG pathway analyses. Many top ranked rules extracted from RANWAR that hold poor ranks in traditional Apriori, are highly biologically significant to the related diseases. Finally, the top rules evolved from RANWAR, that are not in Apriori, are reported.
Including Packages
Our Specialization
Support Service
Statistical Report

satisfied customers
3,589
Freelance projects
983
sales on Site
11,021
developers
175+Additional Information
| Domains | |
|---|---|
| Programming Language |
Would you like to submit yours?















There are no reviews yet