ACID: association correction for imbalanced data in GWAS
Product Description
ACID: association correction for imbalanced data in GWAS
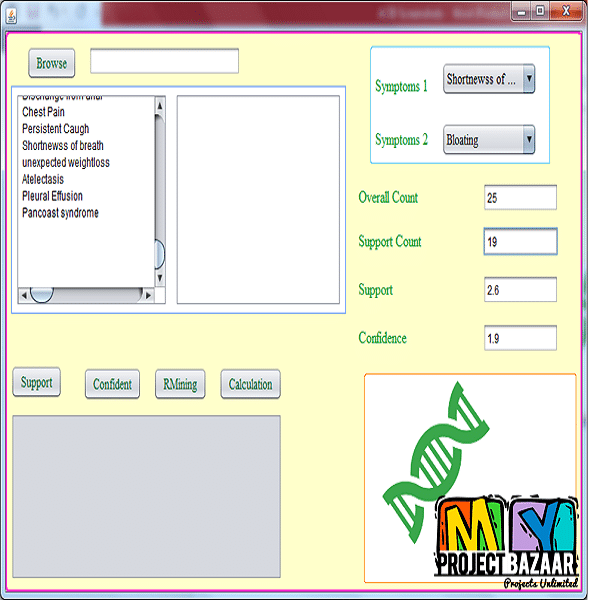
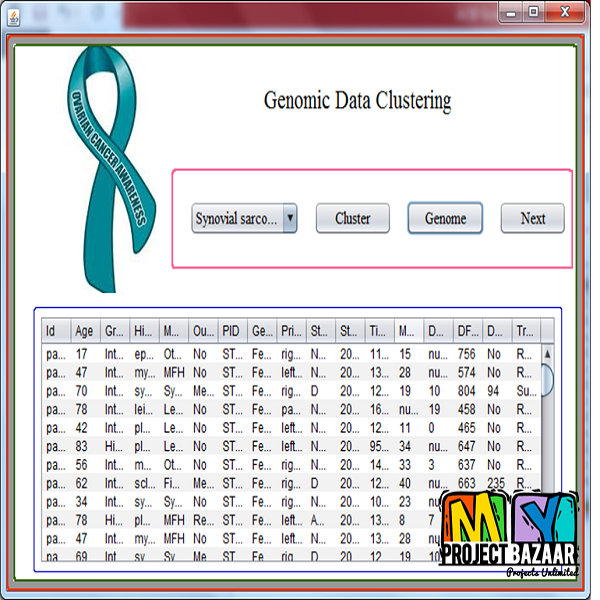
Abstract-Genome-wide association study (GWAS) has been widely witnessed as a powerful tool for revealing suspicious loci from various diseases. However, real world GWAS tasks always suffer from the data imbalance problem of sufficient control samples and limited case samples. This imbalance issue can cause serious biases to the result and thus leads to losses of significance for true causal markers. To tackle this problem, we proposed a computational framework to perform association correction for imbalanced data (ACID) that could potentially improve the performance of GWAS under the imbalance condition. ACID is inspired by the imbalance learning theory but is particularly modified to address the task of association discovery from sequential genomic data. Simulation studies demonstrate ACID can dramatically improve the power of traditional GWAS method on the dataset with severe imbalances. In addition, further applied ACID to two imbalanced datasets (gastric cancer and bladder cancer) to conduct genome wide association analysis. Experimental results indicate that our method has better abilities in identifying suspicious loci than the regression approach and shows consistencies with existing discoveries.
Including Packages
Our Specialization
Support Service
Statistical Report

satisfied customers
3,589
Freelance projects
983
sales on Site
11,021
developers
175+Additional Information
| Domains | |
|---|---|
| Programming Language |