An efficient mixed-model for screening differentially expressed genes of breast cancer based on LR-RF
Product Description
An efficient mixed-model for screening differentially expressed genes of breast cancer based on LR-RF
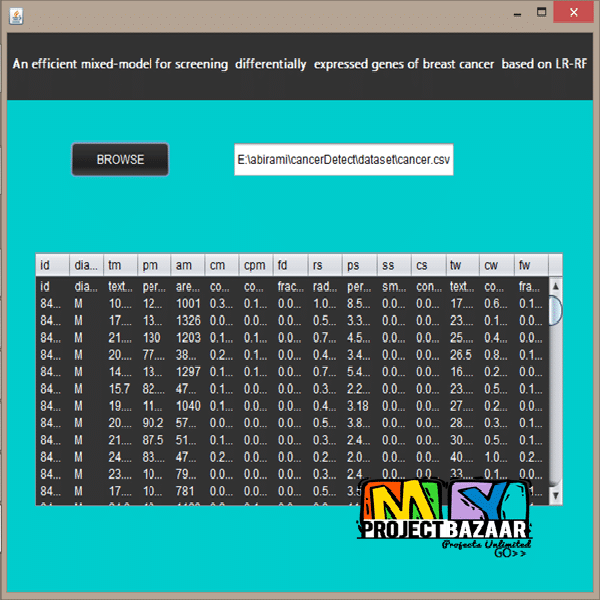
Abstract-To screen differentially expressed genes quickly and efficiently in breast cancer, two gene microarray datasets of breast cancer, GSE15852 and GSE45255, were downloaded from GEO. By combining the Logistic Regression and Random Forest algorithm, this paper proposed a novel method named LR-RF toselect differentially expressed genes of breast cancer on microarray data by the Bonferroni test of FWER error measure. Comparing with Logistic Regression and Random Forest, our study shows that LR-FR has a great facility in selecting differentially expressed genes. The average prediction accuracy of the proposed LR-RF from replicating random test ten times surprisingly reaches 93.11% with variance as low as 0.00045.
The prediction accuracy rate reaches a maximum 95.57% when threshold value 0.2 in the random forest algorithm process of ranking genes’ importance score, and the differentially expressed genes are relatively few in number. In addition, through analyzing the gene interaction networks, most of the top 20 genes we selected were found to involve in the development of breast cancer. All of these results demonstrate the reliability and efficiency of LR-RF. It is anticipated that LR-RF would provide new knowledge and method for biologists, medical scientists, and cognitive computing researchers to identify disease-related genes of breast cancer.
Including Packages
Our Specialization
Support Service
Statistical Report

satisfied customers
3,589
Freelance projects
983
sales on Site
11,021
developers
175+Additional Information
| Domains | |
|---|---|
| Programming Language |